Peter Dazeley
The Bacteria Library: One Hundred Years of Infectious Bugs
Britain’s most important bacterial library, the National Collection of Type Cultures (NCTC) turned 100 this year.
In the century since it was founded in 1920 this biological institution has gathered around 6,000 bacterial strains from more than 900 species, for use in microbiology research worldwide.
Among nearly 800 other registered collections in 78 countries it is the oldest and one of only a few dedicated to bacteria that can make us sick or kill us. Many of the samples contained in the library are reference strains used as test benchmarks by medical institutions.
Scientists use its verified microbial strains to study how bacteria evolve, to test the effects of pesticides or household chemicals, develop vaccines, or anti-cancer drugs and monitor antimicrobial resistance. And when science uncovers new strains – genetic variants – they are deposited in the NCTC so others can study them.
The Origin Story
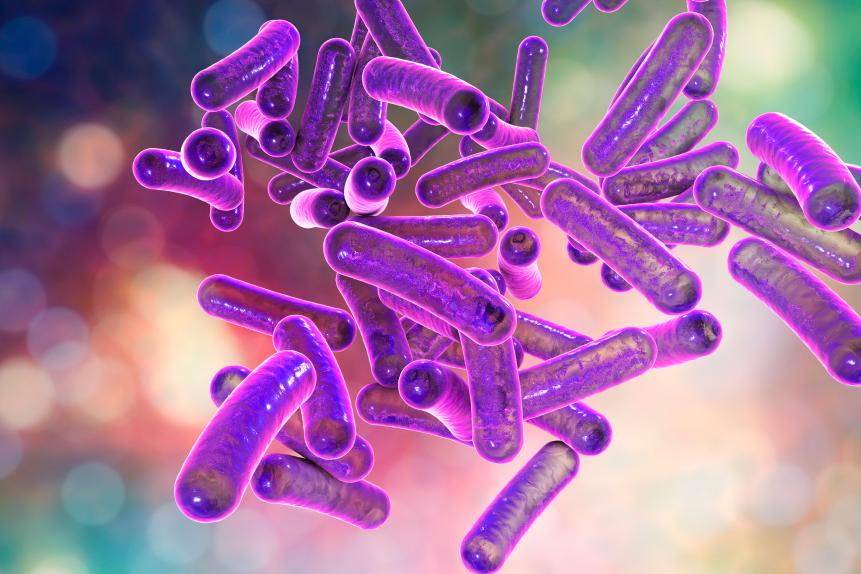
KATERYNA KON/SCIENCE PHOTO LIBRARY
Shigella bacteria, computer illustration. Shigella sp. are Gram-negative rod-shaped bacteria (bacilli) and are the causative agent of human shigellosis. They infect the large intestine and cause dysentery, which can vary in severity from a mild attack of diarrhoea to an acute infection.
The NCTC was established in the aftermath of World War One with a sample of Shigella flexneri, a bacteria causing dysentery and death as it infected soldiers in the trenches. Sir Frederick William Andrewes, a pathologist studying dysentery, deposited S. flexneri (labeled ‘NCTC 1’) from gut bacteria taken from Private Ernest Cable stationed on the Western Front.
In its first year the organization sent out more than 2,000 strains free of charge for research. At the time each sample was delivered alive on a medium made from egg yolks and sealed in paraffin wax. Today new specimens are cultured in agar, then freeze dried and sealed in a glass ampul for long-term storage at 39 degrees Fahrenheit (4 degrees Celsius) – a process similar to one developed in the 1930s.
Antonie van Leeuwenhoek, a Dutch tradesman with an enquiring mind, was the first to identify bacteria way back in the 1670s. Van Leeuwenhoek was recognized by the Royal Society for observing what he termed ‘animalcules’, using microscopes he made himself.
But it wasn’t until physician Alexander Fleming discovered the bacteria-destroying mold that later became penicillin in 1928, that medicine devised effective treatments for microbial infections. Fleming served as a medic during World War One and deposited 16 strains into the NCTC between 1928 and 1948.
Bacteria in War Time
The Great War was a turning point for bacterial science. Strains of the microbe Clostridium perfringens invaded wounds as shells blasted up soil and bodies, causing horrific gas gangrene. Another clostridial disease – tetanus – killed thousands more and knowledge about the impact of bacteria grew quickly.
Improvements in wound care, antiseptics and the introduction of antibiotics greatly reduced bacterial wound infections over the next 20 years. Yet our relationship with bacteria is complex. Clostridium botulinum releases one of the deadliest toxins on Earth, but we use it in cosmetic beauty to eradicate wrinkles.
“More bacteria live and work in one linear centimeter of your lower colon than all the humans who have ever lived. That’s what’s going on in your digestive tract right now. Are we in charge, or are we simply hosts for bacteria?” asked planetary scientist Neil deGrasse Tyson.
Today's Plans for the Germs
Today the NCTC is working to fight what World Health Organization evidence shows is a rapid loss of critically important antimicrobial medicines. Growth in antimicrobial resistance is partly caused by the development of resistant strains, as genes are transferred between bacteria.
Since NCTC1 was first deposited in the bacterial library in 1928, three other strains deposited since then show higher virulence and resistance to antimicrobials. The collection now receives between 50 and 200 strains each year, some as referenced samples, others with their entire genome sequenced.
Over half the collection now has a complete genetic sequence, but the work is ongoing to preserve and update this vital resource. Fighting the infections of the future may rely on it being there in another hundred years.


















